
Dr. Matthias Wielscher, MSc.
Senior Postdoctoral FellowTel.: +43 (0)1 40160-34700
E-Mail: matthias.wielscher@meduniwien.ac.at
Medizinische Universität Wien
Zentrum für Public Health
Abteilung für Epidemiologie
Kinderspitalgasse 15
1090 Wien
Matthias Wielscher
Matthias Wielscher holds a Master of Science (MSc) in Genetics and Microbiology and PhD in Biotechnolgoy. After completing his degree, he spent three years at the Department of Epidemiology and Biostatistics at Imperial College London, working on genome-wide association studies (GWAS) and DNA methylation studies. He then spent two years at Genomics England, contributing to the 100,000 Genomes Project and creating the largest call set from whole genome sequencing data outside the US at that time.
In 2020, Matthias joined the Medical University of Vienna. Since Spring 2023, he has been working with UK Biobank data on the ERC Advanced CLOCKrisk project, using genetic data to understand potential causal relationships between sleep and cardiovascular health and developing genetic scores to predict resilience to night shift work.
Since January 2025, Matthias Wielscher has been focused on his new project titled "Genetic Architecture of Chronic Fatigue Syndrome", which is part of the WWTF Call “ME/CFS 2024 - Understanding ME/CFS”. Currently, there is no consensus on the genetic foundations of ME/CFS, although its heritability estimated at up to 50%. While a large-scale GWAS is ongoing in a dedicated ME/CFS cohort (DECODE-ME), our project will complement this by focusing on rare genetic variants. Thus, the aim of this research project is to provide a list of causal genes for ME/CFS to facilitate further testing and characterization in collaboration with clinical researchers and wet lab biologists.This opportunity arises from the recent availability of whole genome sequencing data from large-scale biobanks such as UK Biobank and All of Us. We can now categorize all protein-altering variants within a gene or other genomic structures like transcription factor binding sites and evaluate their collective impact on ME/CFS. We will utilize the International Consensus Criteria to define cases, however we will also conduct sensitivity analyses around the phenotype definition, and extend our analyses to include long COVID phenotypes within the cohorts mentioned above.
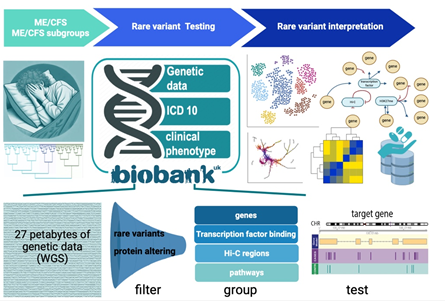
Our primary analysis will be a meta-analysis of data from the UK Biobank and All of Us, which we will, depending on availability, also attempt to replicate using imputed data from the DECODE-ME cohort. This will not only provide the most extensive analysis of rare variants in ME/CFS to date enhancing our knowledge of the condition's causal genes and pathways, but it will also establish a dataspace and collaborative framework for clustering, factor analysis, and other exploratory and epidemiological studies, which we think are essential to fully understand this complex condition. These analyses could also inform subsequent rounds of genetic association tests. Ultimately, the outcomes of these tests may enable stratification of patients and facilitate the categorization of patients into specific subgroups. People involved in the project: Matthias Wielscher (Project Coordinator), Kathryn Hoffman (Co-PI), Golda Schlaff (PostDoc), Leonardo Vincenzi (PostDoc)
